| Previous |
About HAP: Simplicial and cubical complexes |
next |
1. Simplicial Complexes
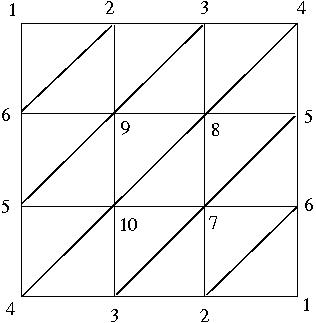
and then calculate its integral homologies from the associated cellular chain complex.
> [5,6,9],[5,9,10],[8,9,10],[7,8,10],[5,7,8],[5,6,7],
> [4,5,10],[3,4,10],[3,7,10],[2,3,7],[2,6,7],[1,2,6]];;
gap> ProjPlane:=MaximalSimplicesToSimplicialComplex(L);
Simplicial complex of dimension 2.
gap> C:=ChainComplex(ProjPlane);
Chain complex of length 2 in characteristic 0 .
gap> Homology(C,0);
[ 0 ]
gap> Homology(C,1);
[ 2 ]
gap> Homology(C,2);
[ ]
gap> n_Sphere:=MaximalSimplicesToSimplicialComplex(Combinations([0..n+1],n+1));
Simplicial complex of dimension 10.
gap> C:=ChainComplex(n_Sphere);
Chain complex of length 10 in characteristic 0 .
gap> List([0..n],m->Homology(C,m));
[ [ 0 ], [ ], [ ], [ ], [ ], [ ], [ ], [ ], [ ], [ ], [ 0 ], [ ] ]
Simplicial complex of dimension 2.
gap> C:=ChainComplex(Q);
Chain complex of length 2 in characteristic 0 .
gap> Homology(C,0);
[ 0 ]
gap> Homology(C,1);
[ ]
gap> Homology(C,2);
[ ]
2. Cubical Complexes
gap> b:=[[1,1,1],[1,0,1],[1,1,1]];;
gap> c:=[[1,1,1],[1,1,1],[1,1,1]];;
gap> array:=[a,b,c];;
gap> 2_sphere:=PureCubicalComplex(array);
Pure cubical complex of dimension 3.
gap> C:=ChainComplex(2_sphere);
Chain complex of length 3 in characteristic 0 .
gap> Homology(C,0);
[ 0 ]
gap> Homology(C,1);
[ ]
gap> Homology(C,2);
[ 0 ]
gap> Homology(C,3);
[ ]
Simplicial complex of dimension 3.
gap> C:=ChainComplex(S);
Chain complex of length 3 in characteristic 0 .
gap> Homology(C,0);
[ 0 ]
gap> Homology(C,1);
[ ]
gap> Homology(C,2);
[ 0 ]
gap> Homology(C,3);
[ ]
Pure cubical complex of dimension 3.
gap> C:=ChainComplex(2_sphere);;
gap> D:=ChainComplex(new_sphere);;
gap> C!.dimension(3);
26
gap> D!.dimension(3);
6
Simplicial complex of dimension 2.
Pure cubical complex of dimension 2.
gap> Torus:=DirectProductOfPureCubicalComplexes(Circle,Circle);
Pure cubical complex of dimension 4.
gap> ContractPureCubicalComplex(Torus);;
gap> SimplicialTorus:=CechComplexOfPureCubicalComplex(Torus);
Simplicial complex of dimension 3.
The following commands use a threshold of 400 to represent the image

as a pure cubical complex. The complex has 40949 2-dimensional cells.
Pure cubical complex of dimension 2.
gap> C:=ChainComplex(image);
Chain complex of length 2 in characteristic 0 .
gap> C!.dimension(0);
45664
gap> C!.dimension(1);
86630
gap> C!.dimension(2);
40949
- Find a homotopy retract R of the pure cubical complex.
- Find a large contractible subcomplex S in R.
- Construct the quotient C(R)/C(S) of the cellular chain complexes.
- Use the fact that Hn(R) = Hn( C(R)/C(S) ) for n>0 and that H0(R) is isomorphic to the direct sum H0(C(R)/C(S))+H0(S).
Pure cubical complex of dimension 2.
gap> R:=ContractedComplex(image);
Pure cubical complex of dimension 2.
gap> S:=ContractibleSubcomplexOfPureCubicalComplex(R);
Pure cubical complex of dimension 2.
gap> C:=ChainComplexOfPair(R,S);
Chain complex of length 2 in characteristic 0 .
gap> Homology(C,0);
[ 0, 0 ]
gap> Homology(C,1);
[ 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 ]
| Previous
Page |
Contents |
Next
page |