Kernel: Python [conda env:py3713]
The (simplified) student Bayes net
This model is from https://github.com/pgmpy/pgmpy_notebook/blob/master/notebooks/2. Bayesian Networks.ipynb
In [1]:
Out[1]:
Collecting pgmpy
Using cached pgmpy-0.1.18-py3-none-any.whl (1.9 MB)
Requirement already satisfied: statsmodels in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (0.13.2)
Requirement already satisfied: scipy in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (1.7.3)
Requirement already satisfied: pandas in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (1.3.5)
Requirement already satisfied: joblib in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (1.1.0)
Requirement already satisfied: tqdm in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (4.64.0)
Requirement already satisfied: pyparsing in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (3.0.9)
Requirement already satisfied: numpy in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (1.21.6)
Requirement already satisfied: scikit-learn in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pgmpy) (1.0.2)
Collecting torch
Downloading torch-1.11.0-cp37-cp37m-manylinux1_x86_64.whl (750.6 MB)
Collecting networkx
Downloading networkx-2.6.3-py3-none-any.whl (1.9 MB)
Requirement already satisfied: python-dateutil>=2.7.3 in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pandas->pgmpy) (2.8.2)
Requirement already satisfied: pytz>=2017.3 in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from pandas->pgmpy) (2022.1)
Requirement already satisfied: six>=1.5 in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from python-dateutil>=2.7.3->pandas->pgmpy) (1.16.0)
Requirement already satisfied: threadpoolctl>=2.0.0 in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from scikit-learn->pgmpy) (3.1.0)
Requirement already satisfied: patsy>=0.5.2 in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from statsmodels->pgmpy) (0.5.2)
Requirement already satisfied: packaging>=21.3 in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from statsmodels->pgmpy) (21.3)
Requirement already satisfied: typing-extensions in /home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages (from torch->pgmpy) (4.2.0)
Installing collected packages: torch, networkx, pgmpy
Successfully installed networkx-2.6.3 pgmpy-0.1.18 torch-1.11.0
In [2]:
Out[2]:
Note: you may need to restart the kernel to use updated packages.
Model
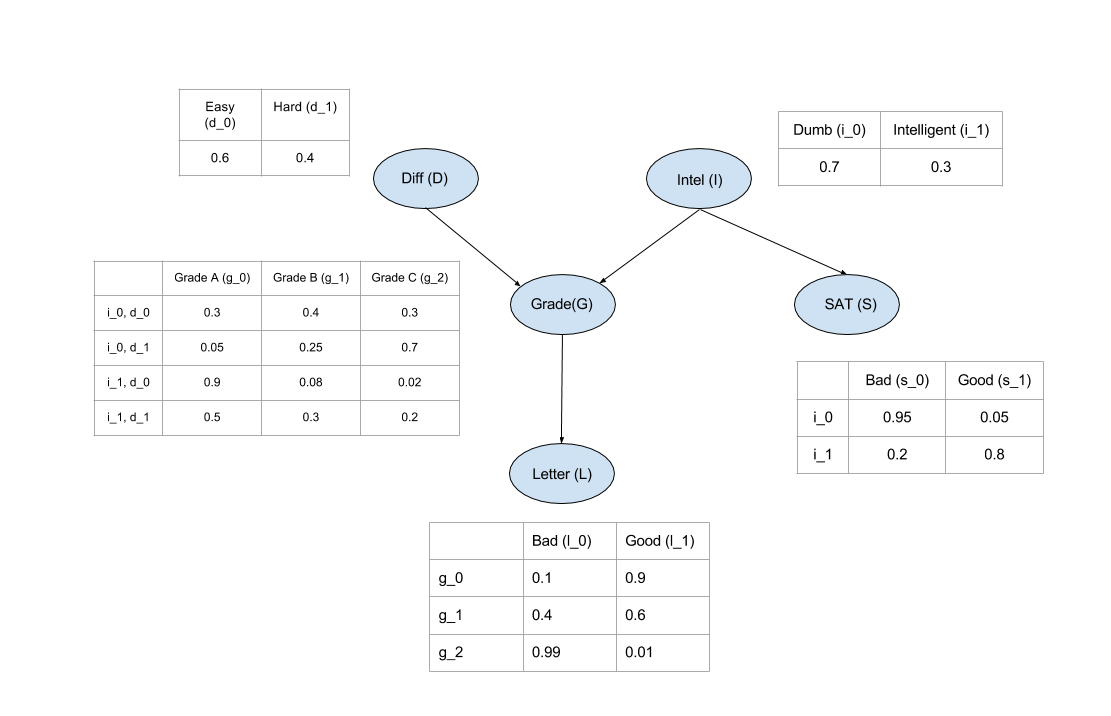
In [3]:
Out[3]:
/home/patel_karm/anaconda3/envs/py3713/lib/python3.7/site-packages/pgmpy/models/BayesianModel.py:10: FutureWarning: BayesianModel has been renamed to BayesianNetwork. Please use BayesianNetwork class, BayesianModel will be removed in future.
FutureWarning,
Basic CPDs
In [4]:
Out[4]:
True
In [5]:
Out[5]:
+----------+----------+----------+----------+----------+
| Intel | Intel(0) | Intel(0) | Intel(1) | Intel(1) |
+----------+----------+----------+----------+----------+
| Diff | Diff(0) | Diff(1) | Diff(0) | Diff(1) |
+----------+----------+----------+----------+----------+
| Grade(0) | 0.3 | 0.05 | 0.9 | 0.5 |
+----------+----------+----------+----------+----------+
| Grade(1) | 0.4 | 0.25 | 0.08 | 0.3 |
+----------+----------+----------+----------+----------+
| Grade(2) | 0.3 | 0.7 | 0.02 | 0.2 |
+----------+----------+----------+----------+----------+
CPDs with names states
In [6]:
Out[6]:
True
In [7]:
Out[7]:
+----------+------------+------------+-------------+-------------+
| Intel | Intel(Low) | Intel(Low) | Intel(High) | Intel(High) |
+----------+------------+------------+-------------+-------------+
| Diff | Diff(Easy) | Diff(Hard) | Diff(Easy) | Diff(Hard) |
+----------+------------+------------+-------------+-------------+
| Grade(A) | 0.3 | 0.05 | 0.9 | 0.5 |
+----------+------------+------------+-------------+-------------+
| Grade(B) | 0.4 | 0.25 | 0.08 | 0.3 |
+----------+------------+------------+-------------+-------------+
| Grade(C) | 0.3 | 0.7 | 0.02 | 0.2 |
+----------+------------+------------+-------------+-------------+
In [8]:
Out[8]:
+------------+-----+
| Diff(Easy) | 0.6 |
+------------+-----+
| Diff(Hard) | 0.4 |
+------------+-----+
+-------------+-----+
| Intel(Low) | 0.7 |
+-------------+-----+
| Intel(High) | 0.3 |
+-------------+-----+
+----------+------------+------------+-------------+-------------+
| Intel | Intel(Low) | Intel(Low) | Intel(High) | Intel(High) |
+----------+------------+------------+-------------+-------------+
| Diff | Diff(Easy) | Diff(Hard) | Diff(Easy) | Diff(Hard) |
+----------+------------+------------+-------------+-------------+
| Grade(A) | 0.3 | 0.05 | 0.9 | 0.5 |
+----------+------------+------------+-------------+-------------+
| Grade(B) | 0.4 | 0.25 | 0.08 | 0.3 |
+----------+------------+------------+-------------+-------------+
| Grade(C) | 0.3 | 0.7 | 0.02 | 0.2 |
+----------+------------+------------+-------------+-------------+
+--------------+----------+----------+----------+
| Grade | Grade(A) | Grade(B) | Grade(C) |
+--------------+----------+----------+----------+
| Letter(Bad) | 0.1 | 0.4 | 0.99 |
+--------------+----------+----------+----------+
| Letter(Good) | 0.9 | 0.6 | 0.01 |
+--------------+----------+----------+----------+
+-----------+------------+-------------+
| Intel | Intel(Low) | Intel(High) |
+-----------+------------+-------------+
| SAT(Bad) | 0.95 | 0.2 |
+-----------+------------+-------------+
| SAT(Good) | 0.05 | 0.8 |
+-----------+------------+-------------+
Inference
In [9]:
Posterior given Grade=C
In [10]:
Out[10]:
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
Pr(Difficulty=Hard|Grade=C) = 0.63
Pr(Intelligence=High|Grade=C) = 0.08
Posterior given Grade=C, SAT=Good
In [11]:
Out[11]:
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
Pr(Difficulty=Hard|Grade=C,SAT=Good) = 0.76
Pr(Intelligence=High|Grade=C,SAT=Good) = 0.58
Visualization
DAG
In [12]:
Out[12]:
<class 'graphviz.graphs.Digraph'>
'student_pgm.pdf'
CPTs
In [14]:
Out[14]:
'student_pgm_with_cpt.pdf'
Marginals
In [15]:
Out[15]:
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0it [00:00, ?it/s]
0it [00:00, ?it/s]
0%| | 0/2 [00:00<?, ?it/s]
0%| | 0/2 [00:00<?, ?it/s]
{'Diff': array([0.37070938, 0.62929062]), 'Grade': array([0., 0., 1.]), 'Intel': array([0.92105263, 0.07894737]), 'Letter': array([0.99, 0.01]), 'SAT': array([0.89078947, 0.10921053])}
In [16]:
Out[16]:
'student_pgm_marginals_given_grade.pdf'
In [17]:
Out[17]:
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0%| | 0/1 [00:00<?, ?it/s]
0it [00:00, ?it/s]
0it [00:00, ?it/s]
{'Diff': array([0.24044002, 0.75955998]), 'Grade': array([0., 0., 1.]), 'Intel': array([0.42168675, 0.57831325]), 'Letter': array([0.99, 0.01]), 'SAT': array([0., 1.])}
'student_pgm_marginals_given_grade_sat.pdf'
In [ ]: