Uncertainty-aware Deep Learning with SNGP
Author: Nimish Sanghi https://github.com/nsanghi
In this notebook we will use JAX, Flax, Optax and Edward2
JAX - JAX is Autograd and XLA, brought together for high-performance numerical computing and machine learning research. It provides composable transformations of Python+NumPy programs: differentiate, vectorize, parallelize, Just-In-Time compile to GPU/TPU, and more.
Flax - Flax is a neural network library and ecosystem for JAX that is designed for flexibility. Flax is in use by a growing community of researchers and engineers at Google who happily use Flax for their daily research.
Optax - Optax is a gradient processing and optimization library for JAX. It is designed to facilitate research by providing building blocks that can be recombined in custom ways in order to optimise parametric models such as, but not limited to, deep neural networks.
Edward2 - Edward2 is a simple probabilistic programming language. It provides core utilities in deep learning ecosystems so that one can write models as probabilistic programs and manipulate a model's computation for flexible training and inference.
This notebook is based on: https://github.com/tensorflow/docs/blob/master/site/en/tutorials/understanding/sngp.ipynb
In AI applications that are safety-critical, such as medical decision making and autonomous driving, or where the data is inherently noisy (for example, natural language understanding), it is important for a deep classifier to reliably quantify its uncertainty. The deep classifier should be able to be aware of its own limitations and when it should hand control over to the human experts. This tutorial shows how to improve a deep classifier's ability in quantifying uncertainty using a technique called Spectral-normalized Neural Gaussian Process SNGP.
The core idea of SNGP is to improve a deep classifier's distance awareness by applying simple modifications to the network. A model's distance awareness is a measure of how its predictive probability reflects the distance between the test example and the training data. This is a desirable property that is common for gold-standard probabilistic models (for example, the Gaussian process with RBF kernels) but is lacking in models with deep neural networks. SNGP provides a simple way to inject this Gaussian-process behavior into a deep classifier while maintaining its predictive accuracy.
This tutorial implements a deep residual network (ResNet)-based SNGP model on scikit-learn’s two moons dataset.
This tutorial illustrates the SNGP model on a toy 2D dataset.
About SNGP
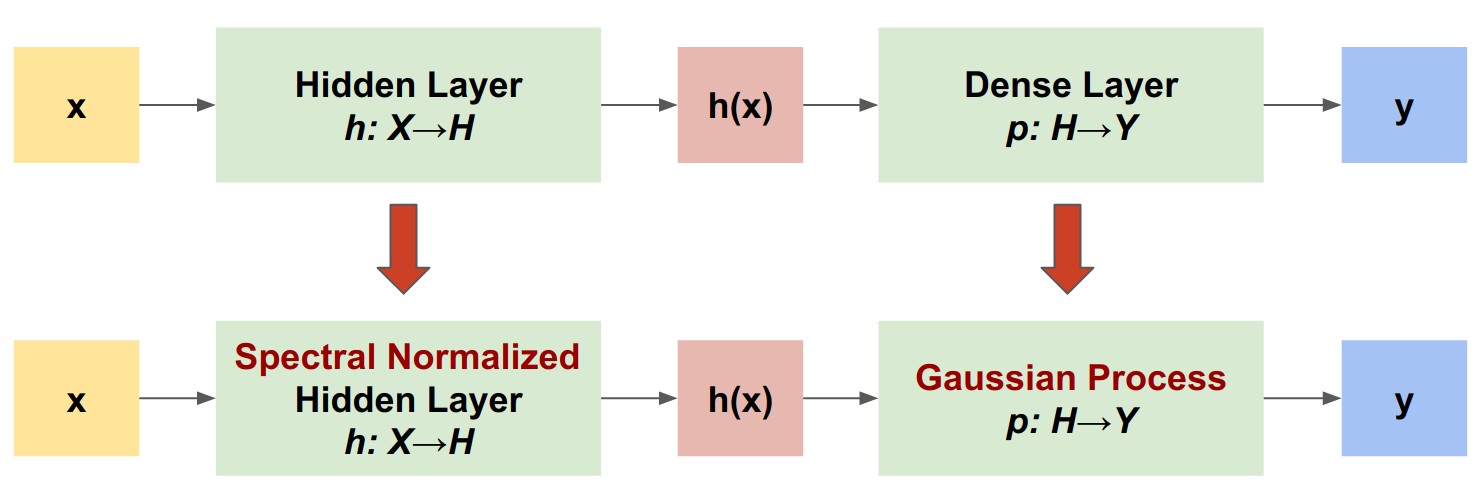
SNGP is a simple approach to improve a deep classifier's uncertainty quality while maintaining a similar level of accuracy and latency. Given a deep residual network, SNGP makes two simple changes to the model:
It applies spectral normalization to the hidden residual layers.
It replaces the Dense output layer with a Gaussian process layer.
Compared to other uncertainty approaches (such as Monte Carlo dropout or Deep ensemble), SNGP has several advantages:
It works for a wide range of state-of-the-art residual-based architectures (for example, (Wide) ResNet, DenseNet, or BERT).
It is a single-model method—it does not rely on ensemble averaging). Therefore, SNGP has a similar level of latency as a single deterministic network, and can be scaled easily to large datasets like ImageNet and Jigsaw Toxic Comments classification
It has strong out-of-domain detection performance due to the distance-awareness property.
The downsides of this method are:
The predictive uncertainty of SNGP is computed using the Laplace approximation. Therefore, theoretically, the posterior uncertainty of SNGP is different from that of an exact Gaussian process.
SNGP training needs a covariance reset step at the beginning of a new epoch. This can add a tiny amount of extra complexity to a training pipeline. This tutorial shows a simple way to implement this using direct update of
stateof the model.
|████████████████████████████████| 180 kB 28.9 MB/s
|████████████████████████████████| 1.0 MB 60.3 MB/s
|████████████████████████████████| 217 kB 69.9 MB/s
|████████████████████████████████| 145 kB 76.2 MB/s
|████████████████████████████████| 51 kB 9.7 MB/s
|████████████████████████████████| 76 kB 7.1 MB/s
Building wheel for jax (setup.py) ... done
Building wheel for edward2 (setup.py) ... done
Define visualization macros
The two moon dataset
Create the training and evaluation datasets from the scikit-learn two moon dataset.
Evaluate the model's predictive behavior over the entire 2D input space.
To evaluate model uncertainty, add an out-of-domain (OOD) dataset that belongs to a third class. The model never observes these OOD examples during training.
Here, the blue and orange represent the positive and negative classes, and the red represents the OOD data. A model that quantifies the uncertainty well is expected to be confident when close to training data (i.e., close to 0 or 1), and be uncertain when far away from the training data regions (i.e., close to 0.5).
The deterministic model
Define model
Start from the (baseline) deterministic model: a multi-layer residual network (ResNet) with dropout regularization.
This tutorial uses a six-layer ResNet with 128 hidden units.
Define Loss and metrics
Create train state
A common pattern in Flax is to create a single dataclass that represents the entire training state, including step number, parameters, and optimizer state.
Train Step
A function that:
Evaluates the neural network given the parameters and a batch of input images with the
Module.applymethod.Computes the
cross_entropy_lossloss function.Evaluates the loss function and its gradient using
jax.value_and_grad.Applies a pytree of gradients to the optimizer to update the model’s parameters.
Computes the metrics using
compute_metrics(defined earlier).
Use JAX’s @jit decorator to trace the entire train_step function and just-in-time compile it with XLA into fused device operations that run faster and more efficiently on hardware accelerators.
Train Function
Initialize the state
Train the model
Visualize uncertainty
Now visualize the predictions of the deterministic model. First plot the class probability:
In this plot, the yellow and purple are the predictive probabilities for the two classes. The deterministic model did a good job in classifying the two known classes—blue and orange—with a nonlinear decision boundary. However, it is not distance-aware, and classified the never-observed red out-of-domain (OOD) examples confidently as the orange class.
Visualize the model uncertainty by computing the predictive variance:
In this plot, the yellow indicates high uncertainty, and the purple indicates low uncertainty. A deterministic ResNet's uncertainty depends only on the test examples' distance from the decision boundary. This leads the model to be over-confident when out of the training domain. The next section shows how SNGP behaves differently on this dataset.
The SNGP model
Define SNGP model
Let's now implement the SNGP model. Both the SNGP components, SpectralNormalization and RandomFeatureGaussianProcess, are available in Edward2.
Let's inspect these two components in more detail.
SpectralNormalization wrapper
SpectralNormalization is a Jax layer wrapper in Edward2 library. It can be applied to an existing Dense layer like this:
Spectral normalization regularizes the hidden weight by gradually guiding its spectral norm (that is, the largest eigenvalue of ) toward the target value norm_multiplier).
Note: Usually it is preferable to set norm_multiplier to a value smaller than 1. However in practice, it can be also relaxed to a larger value to ensure the deep network has enough expressive power.
Next code cell has a simplied implementation of the Spectral Normalization code from Edwards2 library referenced above.
The Gaussian Process (GP) layer
SNGP replaces the typical dense output layer with a Gaussian process (GP) with an RBF kernel, whose posterior variance at is characterized by its distance from the training data in the hidden space.
RandomFeatureGaussianProcess implements a random-feature based approximation to a Gaussian process model that is end-to-end trainable with a deep neural network. Under the hood, the Gaussian process layer implements a two-layer network:
Here, is the input, and and are frozen weights initialized randomly from Gaussian and Uniform distributions, respectively. (Therefore, are called "random features".) is the learnable kernel weight similar to that of a Dense layer.
The main parameters of the GP layers are:
features: The dimension of the output logits.num_inducing: The dimension of the hidden weight . Default to 1024.normalize_input: Whether to apply layer normalization to the input .feature_scale: Defined inhidden_kwargs. UseNoneto apply the scale to the hidden output.momentum: Defined incovmat_kwargs. The momentum of the kernel weight update. Default toNone.
Note: For a deep neural network that is sensitive to the learning rate (for example, ResNet-50 and ResNet-110), it is generally recommended to set normalize_input=True to stabilize training, and set feature_scale=1. to avoid the learning rate from being modified in unexpected ways when passing through the GP layer.
momentumcontrols how the model covariance is computed. If set to a positive value (for example,0.999), the covariance matrix is computed using the momentum-based moving average update (similar to batch normalization). If set toNone, the covariance matrix is updated without momentum.
Note: The momentum-based update method can be sensitive to batch size. Therefore it is generally recommended to set momentum=None to compute the covariance exactly. For this to work properly, the covariance matrix estimator needs to be reset at the beginning of a new epoch in order to avoid counting the same data twice. precision_matrix is the state of the RandomFeatureGaussianProcess layer which we need to access and reset at the begining of each epoch. Function defined below reset_precision_matrix(state) resets the covariance matrix estimator to an Identity matrix.
Given a batch input with shape (batch_size, input_dim), the GP layer returns a logits tensor (shape (batch_size, num_classes)) for prediction, and also covmat tensor (shape (batch_size,) or (batch_size, batch_size)) which is the posterior covariance matrix of the batch logits.
In the code cells below, we implement a simplified version of RandomFeatureGaussianProcess by borrowing the original code from Edwards2 library and removing various configurtaiton settings not relevant to this demonstration as well as hard-coding some of the above recommended settings.
Self Contained Implementation of SNGP layers
RandomFourierFeatures
Code cell implements random features as per equation (6) of the SNGP paper.
The only difference is that as recommended, the code uses a feature_scale of 1 instead of
where, entries in matrix is sampled from and entries in are sampled from . These are sampled in the begining and then fixed. These are not trainable parameters.
LaplaceRandomFeatureCovariance
Notice that under this implementation of the SNGP model as well as in Edwards2 library , the predictive logits for all classes share the same covariance matrix , which describes the distance between from the training data.
Theoretically, it is possible to extend the algorithm to compute different variance values for different classes (as introduced in the original SNGP paper. However, this is difficult to scale to problems with large output spaces (such as classification with ImageNet or language modeling).
RandomFeatureGaussianProcess
Implementation below finally uses the above two functions RandomFourierFeatures and LaplaceRandomFeatureCovariance to implement the distance aware output layer using GP
The hidden features from the last spectral normaized layer in the pipeline are passed through a RandomFoureirFeatures and then a dense layer to get the MAP estimate of logits. These are then passed through LaplaceRandomFeatureCovariance to calculate the posterior covariance of the GP.
The full SNGP model
Given the base class DeepResNet, the SNGP model can be implemented easily by modifying the residual network's hidden and output layers.
Update Step
Next we define update_step which implements a single step of model update. As we need to split the variables into two categories state, the internal state of the model and params, the model's parameters which are updated by the optimizer. state gets updated internally as part of the training step and are required to be extracted and fed back again at the next iteration. And params are the parameters that are updated by the optimizer.
Training over one epoch
THe code below implements a single step through the full training data, shuffling the data at the beginning of each epoch.
Extract precision_matrix from model state and reset
Note: The momentum-based update method can be sensitive to batch size. Therefore it is generally recommended to set momentum=None to compute the covariance exactly. For this to work properly, the covariance matrix estimator needs to be reset at the beginning of a new epoch in order to avoid counting the same data twice.
precision_matrix is the state of the RandomFeatureGaussianProcess layer which we need to access and reset at the begining of each epoch. Function defined below reset_precision_matrix(state) resets the covariance matrix estimator to an Identity matrix.
Init and Train the model
Visualize uncertainty
Next we define the model's prediction and evaluation functions, extracting the probability of the model's predictions. In get_prob_sngp, we first compute the predictive logits and variances.
Next compute the posterior predictive probability. The classic method for computing the predictive probability of a probabilistic model is to use Monte Carlo sampling, i.e.,
where is the sample size, and are random samples from the SNGP posterior (sngp_logits,sngp_covmat). However, this approach can be slow for latency-sensitive applications such as autonomous driving or real-time bidding. Instead, you can approximate using the mean-field method:
where is the SNGP variance, and is often chosen as or .
Note: Instead of fixing to a fixed value, you can also treat it as a hyperparameter, and tune it to optimize the model's calibration performance. This is known as temperature scaling in the deep learning uncertainty literature.
In compute_posterior_mean_probability, we implmention this calculation using the mean-field method.
SNGP Summary
Visualize the class probability (left) and the predictive uncertainty (right) of the SNGP model.
Remember that in the class probability plot (left), the yellow and purple are class probabilities. When close to the training data domain, SNGP correctly classifies the examples with high confidence (i.e., assigning near 0 or 1 probability). When far away from the training data, SNGP gradually becomes less confident, and its predictive probability becomes close to 0.5 while the (normalized) model uncertainty rises to 1.
Compare this to the uncertainty surface of the deterministic model:
As mentioned earlier, a deterministic model is not distance-aware. Its uncertainty is defined by the distance of the test example from the decision boundary. This leads the model to produce overconfident predictions for the out-of-domain examples (red).