%r
library("stringr")
library("reshape2")
library("plyr")
library("ggplot2")
library("MASS")
library("xtable")
if (!file.exists("deaths.rds")) {
deaths <- read.csv("deaths08.csv.bz2")
unlink("deaths08.csv.bz2")
deaths$hod[deaths$hod == 99] <- NA
deaths$hod[deaths$hod == 24] <- 0
deaths$hod[deaths$hod == 0] <- NA
deaths$hod <- as.integer(deaths$hod)
deaths <- arrange(deaths, yod, mod, dod, hod, cod)
deaths <- deaths[c("yod", "mod", "dod", "hod", "cod")]
saveRDS(deaths, "deaths.rds")
}
deaths <- readRDS("deaths.rds")
ok <- subset(deaths, yod == 2008 & mod != 0 & dod != 0)
xtable(ok[c(1, 1:14 * 2000), c("yod", "mod", "dod", "hod", "cod")], "raw.tex")
codes <- read.csv("icd-main.csv")
codes$disease <- sapply(codes$disease, function(x) str_c(strwrap(x, width = 30),collapse = "\n"))
names(codes)[1] <- "cod"
codes <- codes[!duplicated(codes$cod), ]
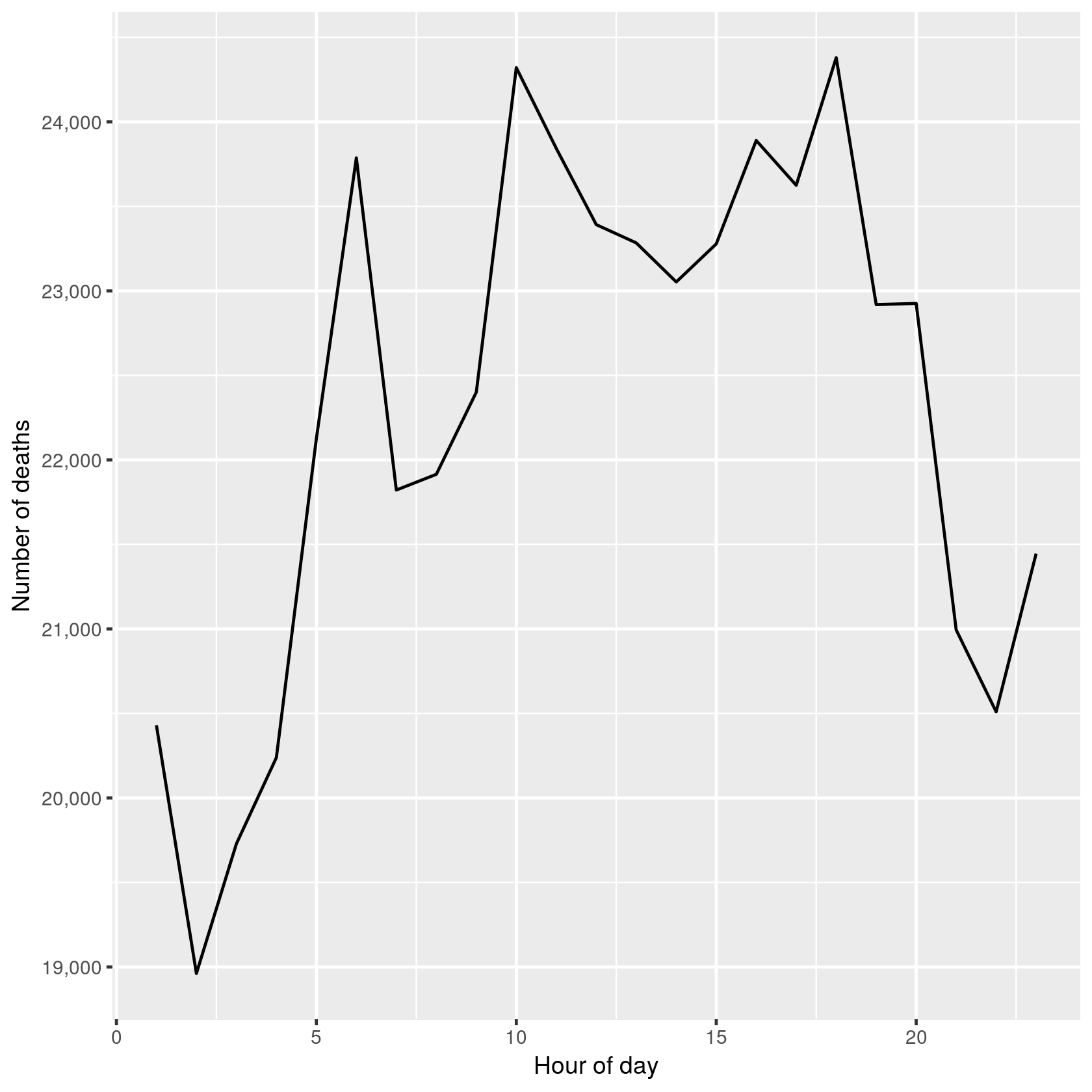
hod_all <- subset(count(deaths, "hod"), !is.na(hod))
qplot(hod, freq, data = hod_all, geom = "line") + scale_y_continuous("Number of deaths",
labels = function(x) format(x, big.mark = ",")) + xlab("Hour of day")